-Search query
-Search result
Showing all 42 items for (author: ciazynska & ka)

EMDB-33748: 
Spike_GSAS_6P protomer RBD domain bound with R1-32 Fab and ACE2 with 3:3:3 ratio
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X
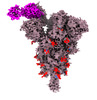
EMDB-33760: 
Spike_GSAS_6P and R1-32 Fab with 3to1 ratio
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X
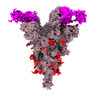
EMDB-33764: 
SARS-COV-2 Spike_GSAS_6P bound with two R1-32 Fabs
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X
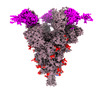
EMDB-33766: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X

EMDB-33772: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X

PDB-7ydi: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2, focused refinement of RBD region
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X

PDB-7ydy: 
SARS-CoV-2 Spike (6P) in complex with 1 R1-32 Fab
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X

PDB-7ye5: 
SARS-CoV-2 Spike (6P) in complex with 2 R1-32 Fabs
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X

PDB-7ye9: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X

PDB-7yeg: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X

EMDB-33453: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-1 Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33454: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-211 Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33455: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-122 Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33456: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33457: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33458: 
Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33459: 
Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33460: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), incubated in Low pH after 40-Day Storage in PBS, Locked-2 Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33461: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site) after 40-Day Storage in PBS, Closed Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33462: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), incubated in Low pH after 40-Day Storage in PBS, Closed Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33463: 
Structure of SARS-CoV-2 D614G spike protein (single Arg S1/S2 cleavage site), Closed Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33464: 
Structure of SARS-Cov-2 D614G spike protein (single Arg S1/S2 cleavage site), Open Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

PDB-7xtz: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-1 Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Zhang X, Carter AP, Xiong X, Briggs JAG

PDB-7xu0: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-211 Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Zhang X, Carter AP, Xiong X, Briggs JAG

PDB-7xu1: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-122 Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Zhang X, Carter AP, Xiong X, Briggs JAG

PDB-7xu2: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Zhang X, Carter AP, Xiong X, Briggs JAG

PDB-7xu3: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Zhang X, Carter AP, Xiong X, Briggs JAG

PDB-7xu4: 
Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Zhang X, Carter AP, Xiong X, Briggs JAG

PDB-7xu5: 
Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Zhang X, Carter AP, Xiong X, Briggs JAG

PDB-7xu6: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), incubated in Low pH after 40-Day Storage in PBS, Locked-2 Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Zhang X, Carter AP, Xiong X, Briggs JAG

EMDB-11329: 
Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x2 disulphide-bond mutant, G413C, V987C, single Arg S1/S2 cleavage site)
Method: single particle / : Qu K, Xiong X, Scheres SHW, Briggs JAG

EMDB-11330: 
Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x1 disulphide-bond mutant, S383C, D985C, K986P, V987P, single Arg S1/S2 cleavage site) in Closed State
Method: single particle / : Qu K, Xiong X, Scheres SHW, Briggs JAG

EMDB-11331: 
Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x1 disulphide-bond mutant, S383C, D985C, K986P, V987P, single Arg S1/S2 cleavage site) in Locked State
Method: single particle / : Qu K, Xiong X, Scheres SHW, Briggs JAG

EMDB-11332: 
Structure of SARS-CoV-2 Spike Protein Trimer (single Arg S1/S2 cleavage site) in Closed State
Method: single particle / : Qu K, Xiong X, Scheres SHW, Briggs JAG

EMDB-11333: 
Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Closed State
Method: single particle / : Qu K, Xiong X, Scheres SHW, Briggs JAG

EMDB-11334: 
Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Locked State
Method: single particle / : Qu K, Xiong X, Scheres SHW, Briggs JAG

PDB-6zox: 
Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x2 disulphide-bond mutant, G413C, V987C, single Arg S1/S2 cleavage site)
Method: single particle / : Xiong X, Qu K, Scheres SHW, Briggs JAG

PDB-6zoy: 
Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x1 disulphide-bond mutant, S383C, D985C, K986P, V987P, single Arg S1/S2 cleavage site) in Closed State
Method: single particle / : Xiong X, Qu K, Scheres SHW, Briggs JAG

PDB-6zoz: 
Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x1 disulphide-bond mutant, S383C, D985C, K986P, V987P, single Arg S1/S2 cleavage site) in Locked State
Method: single particle / : Xiong X, Qu K, Scheres SHW, Briggs JAG

PDB-6zp0: 
Structure of SARS-CoV-2 Spike Protein Trimer (single Arg S1/S2 cleavage site) in Closed State
Method: single particle / : Xiong X, Qu K, Scheres SHW, Briggs JAG

PDB-6zp1: 
Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Closed State
Method: single particle / : Xiong X, Qu K, Scheres SHW, Briggs JAG

PDB-6zp2: 
Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Locked State
Method: single particle / : Xiong X, Qu K, Scheres SHW, Briggs JAG
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
